Glossary¶
Below are definitions of some terms commonly used in the field of pangenomic variation graphs. It is important to note that variation graphs are oriented/directed in pangenomics.
Some definitions can be found from useful websites and publications (see the 'Websites and references' section).
Note that some definitions are specific to certain programs and/or contexts (indicated in square brackets below).
Synopsis¶
Term (French translation) [context if any]: definition.
Terms and definition¶
- Bubble (bulle): Region of the graph representing a variation where multiple paths start and end at common head and tail nodes, respectively. Modified from Eizenga et al. 2020.
- Calling (of variants) (détection/appel de variants) [regular definition]: De novo detection of genetic variants (SNPs or structural variants) by comparison of multiple sequences. SNP calling is often performed by sequence alignment, read mapping to a reference genome or by read stacking (RAD or GBS).
- Calling (of variants) (détection/appel de variants) [Minigraph-Cactus]: Determines which variants in the reads are present in (each haplotype of) the sample. These variants may or may not be present in the graph.
- Genotyping (genotypage) [regular definition]: Allelic/genotypic characterisation of a locus by any possible method.
- Genotyping (genotypage) [graph tools context]: Determines which variants in the graph are present in (each haplotype of) the sample. With NGS, SNP calling and genotyping are now often performed simultaneously by mapping reads to a reference, even for newly considered samples. This is not the case for variation graphs, as one must first build the graph from a discovery panel in order to genotype potential additional samples.
- Graphical Fragment Assembly (GFA): File format storing assembly graphs. It comes in three main versions: rGFA, GFA1 and GFA2 (see gfa-spec repo).
Pangenome variation graph tools mainly use three different flavours:
- Link/Edge(/Relationship) (lien/arête): A connection between two nodes of the graph. Note that in pangenomics, graphs are oriented, so are links (so that they are technically arcs). Links allow overlap between nodes/segments. Links do not carry any other information (e.g. sequence, annotation).
- Node/Segment(/Vertex) (noeud/segment/sommet): Fundamental units forming the graph while connected by edges. In pangenomic variation graphs, nodes correspond to continuous genetic sequences.
- Path (chemin): see 'Walks & paths'.
- Variation graph (graphe de variation): One of the many ways to model a pangenome. Not to be confused with the concept of pangenome. Note that variation graphs in pangenomics are oriented (i.e. directed) and can be cyclic or acyclic (see extended fig. 3 from Liao et al. 2023):
- Cyclic graphs: Graphs that can form loops (walks can pass through the same node and/or edge several times). These graphs are designed to optimise compression. Graphs from PGGB are mainly cyclic, altough they can be built to be locally DAG.
- Directed Acyclic graphs (DAG): Directed graphs that do not form loops (Wikipedia link). The graph is constructed so that haplotype walks don't have to pass the same node or edge twice. Graphs from Minigraph are supposed to be mainly acyclic graphs. This choice make the graph less compressed.
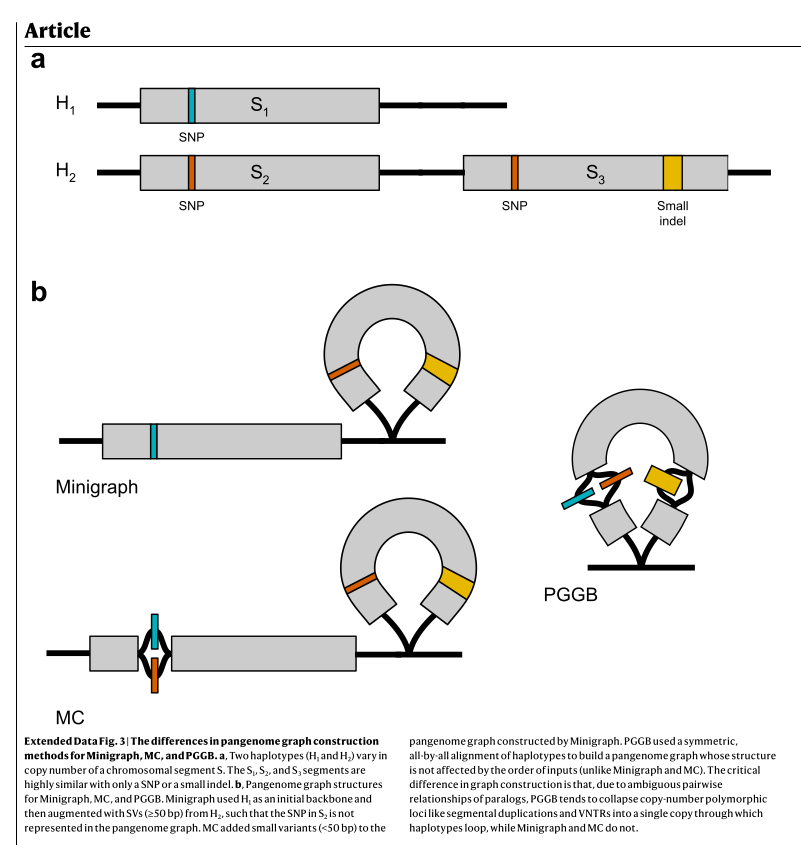 Extended figure 3 from Liao et al. 2023
Extended figure 3 from Liao et al. 2023
- Walks & paths:
- In graph theory, a walk is a sequence of nodes and edges in a graph where you can traverse from one node to another by following the edges.
- In graph theory, a path is a specific type of walk where all nodes (and edges) are distinct. A path does not allow for any repetition of a node or edge.
- In the pangenomic context, only the 'path' concept was defined in the first version of the gfa (1.0) to represent "telomere-to-telomere" sequence and deviated from the graph theory definition by allowing multiple passes through the same node/edge. Path allow overlap between nodes.
- Walks were the defined for gfa1.1 and aim at better representing haplotype sequences in the graph: they does not allow overlap between nodes and can be fragmented.
- Note that walks and paths can exist independently of the sampled haplotypes. However, due to (i) the conceptual proximity of 'walk' and 'path' and (ii) the fact that only 'path' was defined in the first version of the GFA (GFA 1.0) and therefore are more frequently used by pangenomic tools (e.g. visualisation), these terms are often mistakenly used interchangeably.
Websites and references¶
- https://pangenome.github.io/
- Eizenga et al. 2020 (PMC link)
- Liao et al. 2023 (link)
- Matthews et al. 2024 (link)
- The gfa-spec repo
- The rGFA spec repo
- Heng Li's blog (e.g. here and here)